Numbering convention¶
It is important to note that the general numbering convention of features on a chromosome varies depending on the source of the data. For instance, UCSC and Ensembl differ in this point such that an interval labeled (start=4,end=8) will span four base pairs according to UCSC but will span five base pairs according to Ensembl.
A numbering convention for describing continuous intervals of base pairs on a chromosome can be defined by three characteristics:
- Do we start at zero or one ? In track, the smallest authorized number is 0. A feature labeled (start=0,end=4) is legal.
- Are the start and end included ? In track, the start is included and the end is included. A feature labeled (start=X,end=Y) spans Y-X nucleotides.
- Are we counting on the nucleotides or between the nucleotides ? In track, we number between the nucleotides. Two features labeled (start=0,end=4) and (start=4,end=8) have no overlap.
In the following visual examples, we use a feature labeled (start=4,end=8). Both images are considered to be zero-based, start inclusive and end inclusive. However, the image on the left numbers the sugar groups while the image on the right numbers the phosphate groups. This produces different results. This packages sticks to standard depicted on the image labeled “Correct”.
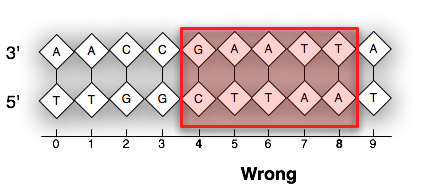
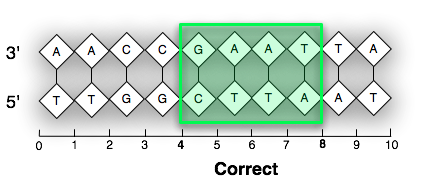
TL;DR: This package uses the UCSC standard for all its representations.